Bio¶
- 👩💻 Software engineer at Bloomberg in NYC
- 🚀 Core developer of numpydoc
- ✍️ Author of Hands-On Data Analysis with Pandas (currently in its second edition; translated into Korean and Simplified Chinese)
- 🎓 Bachelor's degree in operations research from Columbia University
- 🎓 Master's degree in computer science (ML specialization) from Georgia Tech
Prerequisites¶
- To follow along with this workshop, you will need to either configure a local environment or use a cloud solution like GitHub Codespaces. Consult the README for step-by-step setup instructions.
- In addition, you should have basic knowledge of Python and be comfortable working in Jupyter Notebooks; if not, check out the resources here to get up to speed.
Session Outline¶
- Getting Started With Pandas
- Data Wrangling
- Data Visualization
- Hands-On Data Analysis Lab
Section 1: Getting Started With Pandas¶
We will begin by introducing the Series, DataFrame, and Index classes, which are the basic building blocks of the pandas library, and showing how to work with them. By the end of this section, you will be able to create DataFrames and perform operations on them to inspect and filter the data.
Learning Path¶
- Anatomy of a DataFrame
- Creating DataFrames
- Inspecting the data
- Extracting subsets
- Calculating summary statistics
Anatomy of a DataFrame¶
A DataFrame is composed of one or more Series. The names of the Series form the column names, and the row labels form the Index.
import pandas as pd
meteorites = pd.read_csv('../data/Meteorite_Landings.csv', nrows=5)
meteorites
| name | id | nametype | recclass | mass (g) | fall | year | reclat | reclong | GeoLocation | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Aachen | 1 | Valid | L5 | 21 | Fell | 01/01/1880 12:00:00 AM | 50.77500 | 6.08333 | (50.775, 6.08333) |
| 1 | Aarhus | 2 | Valid | H6 | 720 | Fell | 01/01/1951 12:00:00 AM | 56.18333 | 10.23333 | (56.18333, 10.23333) |
| 2 | Abee | 6 | Valid | EH4 | 107000 | Fell | 01/01/1952 12:00:00 AM | 54.21667 | -113.00000 | (54.21667, -113.0) |
| 3 | Acapulco | 10 | Valid | Acapulcoite | 1914 | Fell | 01/01/1976 12:00:00 AM | 16.88333 | -99.90000 | (16.88333, -99.9) |
| 4 | Achiras | 370 | Valid | L6 | 780 | Fell | 01/01/1902 12:00:00 AM | -33.16667 | -64.95000 | (-33.16667, -64.95) |
Source: NASA's Open Data Portal
Series:¶
meteorites.name
0 Aachen 1 Aarhus 2 Abee 3 Acapulco 4 Achiras Name: name, dtype: object
Columns:¶
meteorites.columns
Index(['name', 'id', 'nametype', 'recclass', 'mass (g)', 'fall', 'year',
'reclat', 'reclong', 'GeoLocation'],
dtype='object')
Index:¶
meteorites.index
RangeIndex(start=0, stop=5, step=1)
Learning Path¶
- Anatomy of a DataFrame
- Creating DataFrames
- Inspecting the data
- Extracting subsets
- Calculating summary statistics
Using a flat file¶
import pandas as pd
meteorites = pd.read_csv('../data/Meteorite_Landings.csv')
Using data from an API¶
Collect the data from NASA's Open Data Portal using the Socrata Open Data API (SODA) with the requests library:
import requests
response = requests.get(
'https://data.nasa.gov/docs/legacy/meteorite_landings/gh4g-9sfh.json',
params={'$limit': 50_000}
)
if response.ok:
payload = response.json()
else:
print(f'Request was not successful and returned code: {response.status_code}.')
payload = None
Create the DataFrame with the resulting payload:
import pandas as pd
df = pd.DataFrame(payload)
df.head(3)
| name | id | nametype | recclass | mass | fall | year | reclat | reclong | geolocation | :@computed_region_cbhk_fwbd | :@computed_region_nnqa_25f4 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Aachen | 1 | Valid | L5 | 21 | Fell | 1880-01-01T00:00:00.000 | 50.775000 | 6.083330 | {'latitude': '50.775', 'longitude': '6.08333'} | NaN | NaN |
| 1 | Aarhus | 2 | Valid | H6 | 720 | Fell | 1951-01-01T00:00:00.000 | 56.183330 | 10.233330 | {'latitude': '56.18333', 'longitude': '10.23333'} | NaN | NaN |
| 2 | Abee | 6 | Valid | EH4 | 107000 | Fell | 1952-01-01T00:00:00.000 | 54.216670 | -113.000000 | {'latitude': '54.21667', 'longitude': '-113.0'} | NaN | NaN |
Tip: df.to_csv('data.csv') writes this data to a new file called data.csv.
Learning Path¶
- Anatomy of a DataFrame
- Creating DataFrames
- Inspecting the data
- Extracting subsets
- Calculating summary statistics
Inspecting the data¶
Now that we have some data, we need to perform an initial inspection of it. This gives us information on what the data looks like, how many rows/columns there are, and how much data we have.
Let's inspect the meteorites data.
How many rows and columns are there?¶
meteorites.shape
(45716, 10)
What are the column names?¶
meteorites.columns
Index(['name', 'id', 'nametype', 'recclass', 'mass (g)', 'fall', 'year',
'reclat', 'reclong', 'GeoLocation'],
dtype='object')
What type of data does each column currently hold?¶
meteorites.dtypes
name object id int64 nametype object recclass object mass (g) float64 fall object year object reclat float64 reclong float64 GeoLocation object dtype: object
What does the data look like?¶
meteorites.head()
| name | id | nametype | recclass | mass (g) | fall | year | reclat | reclong | GeoLocation | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Aachen | 1 | Valid | L5 | 21.0 | Fell | 01/01/1880 12:00:00 AM | 50.77500 | 6.08333 | (50.775, 6.08333) |
| 1 | Aarhus | 2 | Valid | H6 | 720.0 | Fell | 01/01/1951 12:00:00 AM | 56.18333 | 10.23333 | (56.18333, 10.23333) |
| 2 | Abee | 6 | Valid | EH4 | 107000.0 | Fell | 01/01/1952 12:00:00 AM | 54.21667 | -113.00000 | (54.21667, -113.0) |
| 3 | Acapulco | 10 | Valid | Acapulcoite | 1914.0 | Fell | 01/01/1976 12:00:00 AM | 16.88333 | -99.90000 | (16.88333, -99.9) |
| 4 | Achiras | 370 | Valid | L6 | 780.0 | Fell | 01/01/1902 12:00:00 AM | -33.16667 | -64.95000 | (-33.16667, -64.95) |
Sometimes there may be extraneous data at the end of the file, so checking the bottom few rows is also important:
meteorites.tail()
| name | id | nametype | recclass | mass (g) | fall | year | reclat | reclong | GeoLocation | |
|---|---|---|---|---|---|---|---|---|---|---|
| 45711 | Zillah 002 | 31356 | Valid | Eucrite | 172.0 | Found | 01/01/1990 12:00:00 AM | 29.03700 | 17.01850 | (29.037, 17.0185) |
| 45712 | Zinder | 30409 | Valid | Pallasite, ungrouped | 46.0 | Found | 01/01/1999 12:00:00 AM | 13.78333 | 8.96667 | (13.78333, 8.96667) |
| 45713 | Zlin | 30410 | Valid | H4 | 3.3 | Found | 01/01/1939 12:00:00 AM | 49.25000 | 17.66667 | (49.25, 17.66667) |
| 45714 | Zubkovsky | 31357 | Valid | L6 | 2167.0 | Found | 01/01/2003 12:00:00 AM | 49.78917 | 41.50460 | (49.78917, 41.5046) |
| 45715 | Zulu Queen | 30414 | Valid | L3.7 | 200.0 | Found | 01/01/1976 12:00:00 AM | 33.98333 | -115.68333 | (33.98333, -115.68333) |
Get some information about the DataFrame¶
meteorites.info()
<class 'pandas.core.frame.DataFrame'> RangeIndex: 45716 entries, 0 to 45715 Data columns (total 10 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 name 45716 non-null object 1 id 45716 non-null int64 2 nametype 45716 non-null object 3 recclass 45716 non-null object 4 mass (g) 45585 non-null float64 5 fall 45716 non-null object 6 year 45425 non-null object 7 reclat 38401 non-null float64 8 reclong 38401 non-null float64 9 GeoLocation 38401 non-null object dtypes: float64(3), int64(1), object(6) memory usage: 3.5+ MB
Solutions¶
Exercise 1.1 – Create a DataFrame by reading in the 2019_Yellow_Taxi_Trip_Data.csv file. Examine the first 5 rows:¶
import pandas as pd
taxis = pd.read_csv('../data/2019_Yellow_Taxi_Trip_Data.csv')
taxis.head()
| vendorid | tpep_pickup_datetime | tpep_dropoff_datetime | passenger_count | trip_distance | ratecodeid | store_and_fwd_flag | pulocationid | dolocationid | payment_type | fare_amount | extra | mta_tax | tip_amount | tolls_amount | improvement_surcharge | total_amount | congestion_surcharge | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2 | 2019-10-23T16:39:42.000 | 2019-10-23T17:14:10.000 | 1 | 7.93 | 1 | N | 138 | 170 | 1 | 29.5 | 1.0 | 0.5 | 7.98 | 6.12 | 0.3 | 47.90 | 2.5 |
| 1 | 1 | 2019-10-23T16:32:08.000 | 2019-10-23T16:45:26.000 | 1 | 2.00 | 1 | N | 11 | 26 | 1 | 10.5 | 1.0 | 0.5 | 0.00 | 0.00 | 0.3 | 12.30 | 0.0 |
| 2 | 2 | 2019-10-23T16:08:44.000 | 2019-10-23T16:21:11.000 | 1 | 1.36 | 1 | N | 163 | 162 | 1 | 9.5 | 1.0 | 0.5 | 2.00 | 0.00 | 0.3 | 15.80 | 2.5 |
| 3 | 2 | 2019-10-23T16:22:44.000 | 2019-10-23T16:43:26.000 | 1 | 1.00 | 1 | N | 170 | 163 | 1 | 13.0 | 1.0 | 0.5 | 4.32 | 0.00 | 0.3 | 21.62 | 2.5 |
| 4 | 2 | 2019-10-23T16:45:11.000 | 2019-10-23T16:58:49.000 | 1 | 1.96 | 1 | N | 163 | 236 | 1 | 10.5 | 1.0 | 0.5 | 0.50 | 0.00 | 0.3 | 15.30 | 2.5 |
Source: NYC Open Data collected via SODA.
Exercise 1.2 – Find the dimensions (number of rows and number of columns) in the data:¶
taxis.shape
(10000, 18)
Learning Path¶
- Anatomy of a DataFrame
- Creating DataFrames
- Inspecting the data
- Extracting subsets
- Calculating summary statistics
Extracting subsets¶
A crucial part of working with DataFrames is extracting subsets of the data: finding rows that meet a certain set of criteria, isolating columns/rows of interest, etc. After narrowing down our data, we are closer to discovering insights. This section will be the backbone of many analysis tasks.
Selecting columns¶
We can select columns as attributes if their names would be valid Python variables:
meteorites.name
0 Aachen
1 Aarhus
2 Abee
3 Acapulco
4 Achiras
...
45711 Zillah 002
45712 Zinder
45713 Zlin
45714 Zubkovsky
45715 Zulu Queen
Name: name, Length: 45716, dtype: object
If they aren't, we have to select them as keys. However, we can select multiple columns at once this way:
meteorites[['name', 'mass (g)']]
| name | mass (g) | |
|---|---|---|
| 0 | Aachen | 21.0 |
| 1 | Aarhus | 720.0 |
| 2 | Abee | 107000.0 |
| 3 | Acapulco | 1914.0 |
| 4 | Achiras | 780.0 |
| ... | ... | ... |
| 45711 | Zillah 002 | 172.0 |
| 45712 | Zinder | 46.0 |
| 45713 | Zlin | 3.3 |
| 45714 | Zubkovsky | 2167.0 |
| 45715 | Zulu Queen | 200.0 |
45716 rows × 2 columns
Selecting rows¶
meteorites[100:104]
| name | id | nametype | recclass | mass (g) | fall | year | reclat | reclong | GeoLocation | |
|---|---|---|---|---|---|---|---|---|---|---|
| 100 | Benton | 5026 | Valid | LL6 | 2840.0 | Fell | 01/01/1949 12:00:00 AM | 45.95000 | -67.55000 | (45.95, -67.55) |
| 101 | Berduc | 48975 | Valid | L6 | 270.0 | Fell | 01/01/2008 12:00:00 AM | -31.91000 | -58.32833 | (-31.91, -58.32833) |
| 102 | Béréba | 5028 | Valid | Eucrite-mmict | 18000.0 | Fell | 01/01/1924 12:00:00 AM | 11.65000 | -3.65000 | (11.65, -3.65) |
| 103 | Berlanguillas | 5029 | Valid | L6 | 1440.0 | Fell | 01/01/1811 12:00:00 AM | 41.68333 | -3.80000 | (41.68333, -3.8) |
Indexing¶
We use iloc[] to select rows and columns by their position:
meteorites.iloc[100:104, [0, 3, 4, 6]]
| name | recclass | mass (g) | year | |
|---|---|---|---|---|
| 100 | Benton | LL6 | 2840.0 | 01/01/1949 12:00:00 AM |
| 101 | Berduc | L6 | 270.0 | 01/01/2008 12:00:00 AM |
| 102 | Béréba | Eucrite-mmict | 18000.0 | 01/01/1924 12:00:00 AM |
| 103 | Berlanguillas | L6 | 1440.0 | 01/01/1811 12:00:00 AM |
We use loc[] to select by name:
meteorites.loc[100:104, 'mass (g)':'year']
| mass (g) | fall | year | |
|---|---|---|---|
| 100 | 2840.0 | Fell | 01/01/1949 12:00:00 AM |
| 101 | 270.0 | Fell | 01/01/2008 12:00:00 AM |
| 102 | 18000.0 | Fell | 01/01/1924 12:00:00 AM |
| 103 | 1440.0 | Fell | 01/01/1811 12:00:00 AM |
| 104 | 960.0 | Fell | 01/01/2004 12:00:00 AM |
Filtering with Boolean masks¶
A Boolean mask is a array-like structure of Boolean values – it's a way to specify which rows/columns we want to select (True) and which we don't (False).
Here's an example of a Boolean mask for meteorites weighing more than 50 grams that were found on Earth (i.e., they were not observed falling):
(meteorites['mass (g)'] > 50) & (meteorites.fall == 'Found')
0 False
1 False
2 False
3 False
4 False
...
45711 True
45712 False
45713 False
45714 True
45715 True
Length: 45716, dtype: bool
Important: Take note of the syntax here. We surround each condition with parentheses, and we use bitwise operators (&, |, ~) instead of logical operators (and, or, not).
We can use a Boolean mask to select the subset of meteorites weighing more than 1 million grams (1,000 kilograms or roughly 2,205 pounds) that were observed falling:
meteorites[(meteorites['mass (g)'] > 1e6) & (meteorites.fall == 'Fell')]
| name | id | nametype | recclass | mass (g) | fall | year | reclat | reclong | GeoLocation | |
|---|---|---|---|---|---|---|---|---|---|---|
| 29 | Allende | 2278 | Valid | CV3 | 2000000.0 | Fell | 01/01/1969 12:00:00 AM | 26.96667 | -105.31667 | (26.96667, -105.31667) |
| 419 | Jilin | 12171 | Valid | H5 | 4000000.0 | Fell | 01/01/1976 12:00:00 AM | 44.05000 | 126.16667 | (44.05, 126.16667) |
| 506 | Kunya-Urgench | 12379 | Valid | H5 | 1100000.0 | Fell | 01/01/1998 12:00:00 AM | 42.25000 | 59.20000 | (42.25, 59.2) |
| 707 | Norton County | 17922 | Valid | Aubrite | 1100000.0 | Fell | 01/01/1948 12:00:00 AM | 39.68333 | -99.86667 | (39.68333, -99.86667) |
| 920 | Sikhote-Alin | 23593 | Valid | Iron, IIAB | 23000000.0 | Fell | 01/01/1947 12:00:00 AM | 46.16000 | 134.65333 | (46.16, 134.65333) |
Tip: Boolean masks can be used with loc[] and iloc[].
An alternative to this is the query() method:
meteorites.query("`mass (g)` > 1e6 and fall == 'Fell'")
| name | id | nametype | recclass | mass (g) | fall | year | reclat | reclong | GeoLocation | |
|---|---|---|---|---|---|---|---|---|---|---|
| 29 | Allende | 2278 | Valid | CV3 | 2000000.0 | Fell | 01/01/1969 12:00:00 AM | 26.96667 | -105.31667 | (26.96667, -105.31667) |
| 419 | Jilin | 12171 | Valid | H5 | 4000000.0 | Fell | 01/01/1976 12:00:00 AM | 44.05000 | 126.16667 | (44.05, 126.16667) |
| 506 | Kunya-Urgench | 12379 | Valid | H5 | 1100000.0 | Fell | 01/01/1998 12:00:00 AM | 42.25000 | 59.20000 | (42.25, 59.2) |
| 707 | Norton County | 17922 | Valid | Aubrite | 1100000.0 | Fell | 01/01/1948 12:00:00 AM | 39.68333 | -99.86667 | (39.68333, -99.86667) |
| 920 | Sikhote-Alin | 23593 | Valid | Iron, IIAB | 23000000.0 | Fell | 01/01/1947 12:00:00 AM | 46.16000 | 134.65333 | (46.16, 134.65333) |
Tip: Here, we can use both logical operators and bitwise operators.
Learning Path¶
- Anatomy of a DataFrame
- Creating DataFrames
- Inspecting the data
- Extracting subsets
- Calculating summary statistics
Calculating summary statistics¶
In the next section of this workshop, we will discuss data cleaning for a more meaningful analysis of our datasets; however, we can already extract some interesting insights from the meteorites data by calculating summary statistics.
How many of the meteorites were found versus observed falling?¶
meteorites.fall.value_counts()
fall Found 44609 Fell 1107 Name: count, dtype: int64
The Meteoritical Society states that a "relict meteorite is dominantly composed of secondary minerals, and is thought to have once been a meteorite." What proportion of the data are relict meteorites? Let's verify these were all found versus observed falling:
meteorites.value_counts(subset=['nametype', 'fall'], normalize=True)
nametype fall
Valid Found 0.974145
Fell 0.024215
Relict Found 0.001641
Name: proportion, dtype: float64
What was the mass of the average meterorite?¶
meteorites['mass (g)'].mean()
np.float64(13278.078548601512)
Important: The mean isn't always the best measure of central tendency. If there are outliers in the distribution, the mean will be skewed. Here, the mean is being pulled higher by some very heavy meteorites – the distribution is right-skewed.
Taking a look at some quantiles at the extremes of the distribution shows that the mean is between the 95th and 99th percentile of the distribution, so it isn't a good measure of central tendency here:
meteorites['mass (g)'].quantile([0.01, 0.05, 0.5, 0.95, 0.99])
0.01 0.44 0.05 1.10 0.50 32.60 0.95 4000.00 0.99 50600.00 Name: mass (g), dtype: float64
A better measure in this case is the median (50th percentile), since it is robust to outliers:
meteorites['mass (g)'].median()
np.float64(32.6)
What was the mass of the heaviest meteorite?¶
meteorites['mass (g)'].max()
np.float64(60000000.0)
Let's extract the information on this meteorite:
meteorites.loc[meteorites['mass (g)'].idxmax()]
name Hoba id 11890 nametype Valid recclass Iron, IVB mass (g) 60000000.0 fall Found year 01/01/1920 12:00:00 AM reclat -19.58333 reclong 17.91667 GeoLocation (-19.58333, 17.91667) Name: 16392, dtype: object
Fun fact: This meteorite landed in Namibia and is a tourist attraction.

How many different types of meteorite classes are represented in this dataset?¶
meteorites.recclass.nunique()
466
Some examples:
meteorites.recclass.unique()[:14]
array(['L5', 'H6', 'EH4', 'Acapulcoite', 'L6', 'LL3-6', 'H5', 'L',
'Diogenite-pm', 'Unknown', 'H4', 'H', 'Iron, IVA', 'CR2-an'],
dtype=object)
Note: All fields preceded with "rec" are the values recommended by The Meteoritical Society. Check out this Wikipedia article for some information on meteorite classes.
Get some summary statistics on the data itself¶
We can get common summary statistics for all columns at once. By default, this will only be numeric columns, but here, we will summarize everything together:
meteorites.describe(include='all')
| name | id | nametype | recclass | mass (g) | fall | year | reclat | reclong | GeoLocation | |
|---|---|---|---|---|---|---|---|---|---|---|
| count | 45716 | 45716.000000 | 45716 | 45716 | 4.558500e+04 | 45716 | 45425 | 38401.000000 | 38401.000000 | 38401 |
| unique | 45716 | NaN | 2 | 466 | NaN | 2 | 266 | NaN | NaN | 17100 |
| top | Aachen | NaN | Valid | L6 | NaN | Found | 01/01/2003 12:00:00 AM | NaN | NaN | (0.0, 0.0) |
| freq | 1 | NaN | 45641 | 8285 | NaN | 44609 | 3323 | NaN | NaN | 6214 |
| mean | NaN | 26889.735104 | NaN | NaN | 1.327808e+04 | NaN | NaN | -39.122580 | 61.074319 | NaN |
| std | NaN | 16860.683030 | NaN | NaN | 5.749889e+05 | NaN | NaN | 46.378511 | 80.647298 | NaN |
| min | NaN | 1.000000 | NaN | NaN | 0.000000e+00 | NaN | NaN | -87.366670 | -165.433330 | NaN |
| 25% | NaN | 12688.750000 | NaN | NaN | 7.200000e+00 | NaN | NaN | -76.714240 | 0.000000 | NaN |
| 50% | NaN | 24261.500000 | NaN | NaN | 3.260000e+01 | NaN | NaN | -71.500000 | 35.666670 | NaN |
| 75% | NaN | 40656.750000 | NaN | NaN | 2.026000e+02 | NaN | NaN | 0.000000 | 157.166670 | NaN |
| max | NaN | 57458.000000 | NaN | NaN | 6.000000e+07 | NaN | NaN | 81.166670 | 354.473330 | NaN |
Important: NaN values signify missing data. For instance, the fall column contains strings, so there is no value for mean; likewise, mass (g) is numeric, so we don't have entries for the categorical summary statistics (unique, top, freq).
Exercise 1.3¶
Using the data in the 2019_Yellow_Taxi_Trip_Data.csv file, calculate summary statistics for the fare_amount, tip_amount, tolls_amount, and total_amount columns.¶
Exercise 1.4¶
Isolate the fare_amount, tip_amount, tolls_amount, and total_amount for the longest trip by distance (trip_distance).¶
Solutions¶
Exercise 1.3 – Using the data in the 2019_Yellow_Taxi_Trip_Data.csv file, calculate summary statistics for the fare_amount, tip_amount, tolls_amount, and total_amount columns:¶
import pandas as pd
taxis = pd.read_csv('../data/2019_Yellow_Taxi_Trip_Data.csv')
taxis[['fare_amount', 'tip_amount', 'tolls_amount', 'total_amount']].describe()
| fare_amount | tip_amount | tolls_amount | total_amount | |
|---|---|---|---|---|
| count | 10000.000000 | 10000.000000 | 10000.000000 | 10000.000000 |
| mean | 15.106313 | 2.634494 | 0.623447 | 22.564659 |
| std | 13.954762 | 3.409800 | 6.437507 | 19.209255 |
| min | -52.000000 | 0.000000 | -6.120000 | -65.920000 |
| 25% | 7.000000 | 0.000000 | 0.000000 | 12.375000 |
| 50% | 10.000000 | 2.000000 | 0.000000 | 16.300000 |
| 75% | 16.000000 | 3.250000 | 0.000000 | 22.880000 |
| max | 176.000000 | 43.000000 | 612.000000 | 671.800000 |
Exercise 1.4 – Isolate the fare_amount, tip_amount, tolls_amount, and total_amount for the longest trip by distance (trip_distance):¶
taxis.loc[
taxis.trip_distance.idxmax(),
['fare_amount', 'tip_amount', 'tolls_amount', 'total_amount']
]
fare_amount 176.0 tip_amount 18.29 tolls_amount 6.12 total_amount 201.21 Name: 8338, dtype: object
Section 1 Complete 🎉¶

Section 2: Data Wrangling¶
To prepare our data for analysis, we need to perform data wrangling. In this section, we will learn how to clean and reformat data (e.g., renaming columns and fixing data type mismatches), restructure/reshape it, and enrich it (e.g., discretizing columns, calculating aggregations, and combining data sources).
Learning Path¶
- Data cleaning
- Working with the index
- Reshaping data
- Aggregations and grouping
- Time series
Data cleaning¶
In this section, we will take a look at creating, renaming, and dropping columns; type conversion; and sorting – all of which make our analysis easier. We will be working with the 2019 Yellow Taxi Trip Data provided by NYC Open Data.
import pandas as pd
taxis = pd.read_csv('../data/2019_Yellow_Taxi_Trip_Data.csv')
taxis.head()
| vendorid | tpep_pickup_datetime | tpep_dropoff_datetime | passenger_count | trip_distance | ratecodeid | store_and_fwd_flag | pulocationid | dolocationid | payment_type | fare_amount | extra | mta_tax | tip_amount | tolls_amount | improvement_surcharge | total_amount | congestion_surcharge | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2 | 2019-10-23T16:39:42.000 | 2019-10-23T17:14:10.000 | 1 | 7.93 | 1 | N | 138 | 170 | 1 | 29.5 | 1.0 | 0.5 | 7.98 | 6.12 | 0.3 | 47.90 | 2.5 |
| 1 | 1 | 2019-10-23T16:32:08.000 | 2019-10-23T16:45:26.000 | 1 | 2.00 | 1 | N | 11 | 26 | 1 | 10.5 | 1.0 | 0.5 | 0.00 | 0.00 | 0.3 | 12.30 | 0.0 |
| 2 | 2 | 2019-10-23T16:08:44.000 | 2019-10-23T16:21:11.000 | 1 | 1.36 | 1 | N | 163 | 162 | 1 | 9.5 | 1.0 | 0.5 | 2.00 | 0.00 | 0.3 | 15.80 | 2.5 |
| 3 | 2 | 2019-10-23T16:22:44.000 | 2019-10-23T16:43:26.000 | 1 | 1.00 | 1 | N | 170 | 163 | 1 | 13.0 | 1.0 | 0.5 | 4.32 | 0.00 | 0.3 | 21.62 | 2.5 |
| 4 | 2 | 2019-10-23T16:45:11.000 | 2019-10-23T16:58:49.000 | 1 | 1.96 | 1 | N | 163 | 236 | 1 | 10.5 | 1.0 | 0.5 | 0.50 | 0.00 | 0.3 | 15.30 | 2.5 |
Source: NYC Open Data collected via SODA.
Dropping columns¶
Let's start by dropping the ID columns and the store_and_fwd_flag column, which we won't be using.
mask = taxis.columns.str.contains('id$|store_and_fwd_flag', regex=True)
columns_to_drop = taxis.columns[mask]
columns_to_drop
Index(['vendorid', 'ratecodeid', 'store_and_fwd_flag', 'pulocationid',
'dolocationid'],
dtype='object')
taxis = taxis.drop(columns=columns_to_drop)
taxis.head()
| tpep_pickup_datetime | tpep_dropoff_datetime | passenger_count | trip_distance | payment_type | fare_amount | extra | mta_tax | tip_amount | tolls_amount | improvement_surcharge | total_amount | congestion_surcharge | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2019-10-23T16:39:42.000 | 2019-10-23T17:14:10.000 | 1 | 7.93 | 1 | 29.5 | 1.0 | 0.5 | 7.98 | 6.12 | 0.3 | 47.90 | 2.5 |
| 1 | 2019-10-23T16:32:08.000 | 2019-10-23T16:45:26.000 | 1 | 2.00 | 1 | 10.5 | 1.0 | 0.5 | 0.00 | 0.00 | 0.3 | 12.30 | 0.0 |
| 2 | 2019-10-23T16:08:44.000 | 2019-10-23T16:21:11.000 | 1 | 1.36 | 1 | 9.5 | 1.0 | 0.5 | 2.00 | 0.00 | 0.3 | 15.80 | 2.5 |
| 3 | 2019-10-23T16:22:44.000 | 2019-10-23T16:43:26.000 | 1 | 1.00 | 1 | 13.0 | 1.0 | 0.5 | 4.32 | 0.00 | 0.3 | 21.62 | 2.5 |
| 4 | 2019-10-23T16:45:11.000 | 2019-10-23T16:58:49.000 | 1 | 1.96 | 1 | 10.5 | 1.0 | 0.5 | 0.50 | 0.00 | 0.3 | 15.30 | 2.5 |
Tip: Another way to do this is to select the columns we want to keep: taxis.loc[:,~mask].
Renaming columns¶
Next, let's rename the datetime columns:
taxis = taxis.rename(
columns={
'tpep_pickup_datetime': 'pickup',
'tpep_dropoff_datetime': 'dropoff'
}
)
taxis.columns
Index(['pickup', 'dropoff', 'passenger_count', 'trip_distance', 'payment_type',
'fare_amount', 'extra', 'mta_tax', 'tip_amount', 'tolls_amount',
'improvement_surcharge', 'total_amount', 'congestion_surcharge'],
dtype='object')
Type conversion¶
Notice anything off with the data types?
taxis.dtypes
pickup object dropoff object passenger_count int64 trip_distance float64 payment_type int64 fare_amount float64 extra float64 mta_tax float64 tip_amount float64 tolls_amount float64 improvement_surcharge float64 total_amount float64 congestion_surcharge float64 dtype: object
Both pickup and dropoff should be stored as datetimes. Let's fix this:
taxis[['pickup', 'dropoff']] = \
taxis[['pickup', 'dropoff']].apply(pd.to_datetime)
taxis.dtypes
pickup datetime64[ns] dropoff datetime64[ns] passenger_count int64 trip_distance float64 payment_type int64 fare_amount float64 extra float64 mta_tax float64 tip_amount float64 tolls_amount float64 improvement_surcharge float64 total_amount float64 congestion_surcharge float64 dtype: object
Tip: There are other ways to perform type conversion. For numeric values, we can use the pd.to_numeric() function, and we will see the astype() method, which is a more generic method, a little later.
Creating new columns¶
Let's calculate the following for each row:
- elapsed time of the trip
- the tip percentage
- the total taxes, tolls, fees, and surcharges
- the average speed of the taxi
taxis = taxis.assign(
elapsed_time=lambda x: x.dropoff - x.pickup, # 1
cost_before_tip=lambda x: x.total_amount - x.tip_amount,
tip_pct=lambda x: x.tip_amount / x.cost_before_tip, # 2
fees=lambda x: x.cost_before_tip - x.fare_amount, # 3
avg_speed=lambda x: x.trip_distance.div(
x.elapsed_time.dt.total_seconds() / 60 / 60
) # 4
)
Tip: New to lambda functions? These small, anonymous functions can receive multiple arguments, but can only contain one expression (the return value). You will see these a lot in pandas code. Read more about them here.
Our new columns get added to the right:
taxis.head(2)
| pickup | dropoff | passenger_count | trip_distance | payment_type | fare_amount | extra | mta_tax | tip_amount | tolls_amount | improvement_surcharge | total_amount | congestion_surcharge | elapsed_time | cost_before_tip | tip_pct | fees | avg_speed | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2019-10-23 16:39:42 | 2019-10-23 17:14:10 | 1 | 7.93 | 1 | 29.5 | 1.0 | 0.5 | 7.98 | 6.12 | 0.3 | 47.9 | 2.5 | 0 days 00:34:28 | 39.92 | 0.1999 | 10.42 | 13.804642 |
| 1 | 2019-10-23 16:32:08 | 2019-10-23 16:45:26 | 1 | 2.00 | 1 | 10.5 | 1.0 | 0.5 | 0.00 | 0.00 | 0.3 | 12.3 | 0.0 | 0 days 00:13:18 | 12.30 | 0.0000 | 1.80 | 9.022556 |
Some things to note:
- We used
lambdafunctions to 1) avoid typingtaxisrepeatedly and 2) be able to access thecost_before_tipandelapsed_timecolumns in the same method that we create them. - To create a single new column, we can also use
df['new_col'] = <values>.
Sorting by values¶
We can use the sort_values() method to sort based on any number of columns:
taxis.sort_values(['passenger_count', 'pickup'], ascending=[False, True]).head()
| pickup | dropoff | passenger_count | trip_distance | payment_type | fare_amount | extra | mta_tax | tip_amount | tolls_amount | improvement_surcharge | total_amount | congestion_surcharge | elapsed_time | cost_before_tip | tip_pct | fees | avg_speed | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5997 | 2019-10-23 15:55:19 | 2019-10-23 16:08:25 | 6 | 1.58 | 2 | 10.0 | 1.0 | 0.5 | 0.0 | 0.0 | 0.3 | 14.3 | 2.5 | 0 days 00:13:06 | 14.3 | 0.000000 | 4.3 | 7.236641 |
| 443 | 2019-10-23 15:56:59 | 2019-10-23 16:04:33 | 6 | 1.46 | 2 | 7.5 | 1.0 | 0.5 | 0.0 | 0.0 | 0.3 | 11.8 | 2.5 | 0 days 00:07:34 | 11.8 | 0.000000 | 4.3 | 11.577093 |
| 8722 | 2019-10-23 15:57:33 | 2019-10-23 16:03:34 | 6 | 0.62 | 1 | 5.5 | 1.0 | 0.5 | 0.7 | 0.0 | 0.3 | 10.5 | 2.5 | 0 days 00:06:01 | 9.8 | 0.071429 | 4.3 | 6.182825 |
| 4198 | 2019-10-23 15:57:38 | 2019-10-23 16:05:07 | 6 | 1.18 | 1 | 7.0 | 1.0 | 0.5 | 1.0 | 0.0 | 0.3 | 12.3 | 2.5 | 0 days 00:07:29 | 11.3 | 0.088496 | 4.3 | 9.461024 |
| 8238 | 2019-10-23 15:58:31 | 2019-10-23 16:29:29 | 6 | 3.23 | 2 | 19.5 | 1.0 | 0.5 | 0.0 | 0.0 | 0.3 | 23.8 | 2.5 | 0 days 00:30:58 | 23.8 | 0.000000 | 4.3 | 6.258342 |
To pick out the largest/smallest rows, use nlargest() / nsmallest() instead. Looking at the 3 trips with the longest elapsed time, we see some possible data integrity issues:
taxis.nlargest(3, 'elapsed_time')
| pickup | dropoff | passenger_count | trip_distance | payment_type | fare_amount | extra | mta_tax | tip_amount | tolls_amount | improvement_surcharge | total_amount | congestion_surcharge | elapsed_time | cost_before_tip | tip_pct | fees | avg_speed | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 7576 | 2019-10-23 16:52:51 | 2019-10-24 16:51:44 | 1 | 3.75 | 1 | 17.5 | 1.0 | 0.5 | 0.0 | 0.0 | 0.3 | 21.8 | 2.5 | 0 days 23:58:53 | 21.8 | 0.0 | 4.3 | 0.156371 |
| 6902 | 2019-10-23 16:51:42 | 2019-10-24 16:50:22 | 1 | 11.19 | 2 | 39.5 | 1.0 | 0.5 | 0.0 | 0.0 | 0.3 | 41.3 | 0.0 | 0 days 23:58:40 | 41.3 | 0.0 | 1.8 | 0.466682 |
| 4975 | 2019-10-23 16:18:51 | 2019-10-24 16:17:30 | 1 | 0.70 | 2 | 7.0 | 1.0 | 0.5 | 0.0 | 0.0 | 0.3 | 11.3 | 2.5 | 0 days 23:58:39 | 11.3 | 0.0 | 4.3 | 0.029194 |
Solution¶
import pandas as pd
meteorites = pd.read_csv('../data/Meteorite_Landings.csv')
meteorites = meteorites\
.rename(columns={'mass (g)': 'mass'})\
.drop(columns=meteorites.columns[-3:])\
.sort_values('mass', ascending=False)
meteorites.head()
| name | id | nametype | recclass | mass | fall | year | |
|---|---|---|---|---|---|---|---|
| 16392 | Hoba | 11890 | Valid | Iron, IVB | 60000000.0 | Found | 01/01/1920 12:00:00 AM |
| 5373 | Cape York | 5262 | Valid | Iron, IIIAB | 58200000.0 | Found | 01/01/1818 12:00:00 AM |
| 5365 | Campo del Cielo | 5247 | Valid | Iron, IAB-MG | 50000000.0 | Found | 12/22/1575 12:00:00 AM |
| 5370 | Canyon Diablo | 5257 | Valid | Iron, IAB-MG | 30000000.0 | Found | 01/01/1891 12:00:00 AM |
| 3455 | Armanty | 2335 | Valid | Iron, IIIE | 28000000.0 | Found | 01/01/1898 12:00:00 AM |
Learning Path¶
- Data cleaning
- Working with the index
- Reshaping data
- Aggregations and grouping
- Time series
Working with the index¶
So far, we haven't really worked with the index because it's just been a row number; however, we can change the values we have in the index to access additional features of the pandas library.
Setting and sorting the index¶
Currently, we have a RangeIndex, but we can switch to a DatetimeIndex by specifying a datetime column when calling set_index():
taxis = taxis.set_index('pickup')
taxis.head(3)
| dropoff | passenger_count | trip_distance | payment_type | fare_amount | extra | mta_tax | tip_amount | tolls_amount | improvement_surcharge | total_amount | congestion_surcharge | elapsed_time | cost_before_tip | tip_pct | fees | avg_speed | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pickup | |||||||||||||||||
| 2019-10-23 16:39:42 | 2019-10-23 17:14:10 | 1 | 7.93 | 1 | 29.5 | 1.0 | 0.5 | 7.98 | 6.12 | 0.3 | 47.9 | 2.5 | 0 days 00:34:28 | 39.92 | 0.199900 | 10.42 | 13.804642 |
| 2019-10-23 16:32:08 | 2019-10-23 16:45:26 | 1 | 2.00 | 1 | 10.5 | 1.0 | 0.5 | 0.00 | 0.00 | 0.3 | 12.3 | 0.0 | 0 days 00:13:18 | 12.30 | 0.000000 | 1.80 | 9.022556 |
| 2019-10-23 16:08:44 | 2019-10-23 16:21:11 | 1 | 1.36 | 1 | 9.5 | 1.0 | 0.5 | 2.00 | 0.00 | 0.3 | 15.8 | 2.5 | 0 days 00:12:27 | 13.80 | 0.144928 | 4.30 | 6.554217 |
Since we have a sample of the full dataset, let's sort the index to order by pickup time:
taxis = taxis.sort_index()
Tip: taxis.sort_index(axis=1) will sort the columns by name. The axis parameter is present throughout the pandas library: axis=0 targets rows and axis=1 targets columns.
We can now select ranges from our data based on the datetime the same way we did with row numbers:
taxis['2019-10-23 07:45':'2019-10-23 08']
| dropoff | passenger_count | trip_distance | payment_type | fare_amount | extra | mta_tax | tip_amount | tolls_amount | improvement_surcharge | total_amount | congestion_surcharge | elapsed_time | cost_before_tip | tip_pct | fees | avg_speed | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pickup | |||||||||||||||||
| 2019-10-23 07:48:58 | 2019-10-23 07:52:09 | 1 | 0.67 | 2 | 4.5 | 1.0 | 0.5 | 0.0 | 0.0 | 0.3 | 8.8 | 2.5 | 0 days 00:03:11 | 8.8 | 0.000000 | 4.3 | 12.628272 |
| 2019-10-23 08:02:09 | 2019-10-24 07:42:32 | 1 | 8.38 | 1 | 32.0 | 1.0 | 0.5 | 5.5 | 0.0 | 0.3 | 41.8 | 2.5 | 0 days 23:40:23 | 36.3 | 0.151515 | 4.3 | 0.353989 |
| 2019-10-23 08:18:47 | 2019-10-23 08:36:05 | 1 | 2.39 | 2 | 12.5 | 1.0 | 0.5 | 0.0 | 0.0 | 0.3 | 16.8 | 2.5 | 0 days 00:17:18 | 16.8 | 0.000000 | 4.3 | 8.289017 |
When not specifying a range, we use loc[]:
taxis.loc['2019-10-23 08']
| dropoff | passenger_count | trip_distance | payment_type | fare_amount | extra | mta_tax | tip_amount | tolls_amount | improvement_surcharge | total_amount | congestion_surcharge | elapsed_time | cost_before_tip | tip_pct | fees | avg_speed | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pickup | |||||||||||||||||
| 2019-10-23 08:02:09 | 2019-10-24 07:42:32 | 1 | 8.38 | 1 | 32.0 | 1.0 | 0.5 | 5.5 | 0.0 | 0.3 | 41.8 | 2.5 | 0 days 23:40:23 | 36.3 | 0.151515 | 4.3 | 0.353989 |
| 2019-10-23 08:18:47 | 2019-10-23 08:36:05 | 1 | 2.39 | 2 | 12.5 | 1.0 | 0.5 | 0.0 | 0.0 | 0.3 | 16.8 | 2.5 | 0 days 00:17:18 | 16.8 | 0.000000 | 4.3 | 8.289017 |
Resetting the index¶
We will be working with time series later this section, but sometimes we want to reset our index to row numbers and restore the columns. We can make pickup a column again with the reset_index() method:
taxis = taxis.reset_index()
taxis.head()
| pickup | dropoff | passenger_count | trip_distance | payment_type | fare_amount | extra | mta_tax | tip_amount | tolls_amount | improvement_surcharge | total_amount | congestion_surcharge | elapsed_time | cost_before_tip | tip_pct | fees | avg_speed | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2019-10-23 07:05:34 | 2019-10-23 08:03:16 | 3 | 14.68 | 1 | 50.0 | 1.0 | 0.5 | 4.0 | 0.0 | 0.3 | 55.8 | 0.0 | 0 days 00:57:42 | 51.8 | 0.077220 | 1.8 | 15.265165 |
| 1 | 2019-10-23 07:48:58 | 2019-10-23 07:52:09 | 1 | 0.67 | 2 | 4.5 | 1.0 | 0.5 | 0.0 | 0.0 | 0.3 | 8.8 | 2.5 | 0 days 00:03:11 | 8.8 | 0.000000 | 4.3 | 12.628272 |
| 2 | 2019-10-23 08:02:09 | 2019-10-24 07:42:32 | 1 | 8.38 | 1 | 32.0 | 1.0 | 0.5 | 5.5 | 0.0 | 0.3 | 41.8 | 2.5 | 0 days 23:40:23 | 36.3 | 0.151515 | 4.3 | 0.353989 |
| 3 | 2019-10-23 08:18:47 | 2019-10-23 08:36:05 | 1 | 2.39 | 2 | 12.5 | 1.0 | 0.5 | 0.0 | 0.0 | 0.3 | 16.8 | 2.5 | 0 days 00:17:18 | 16.8 | 0.000000 | 4.3 | 8.289017 |
| 4 | 2019-10-23 09:27:16 | 2019-10-23 09:33:13 | 2 | 1.11 | 2 | 6.0 | 1.0 | 0.5 | 0.0 | 0.0 | 0.3 | 7.8 | 0.0 | 0 days 00:05:57 | 7.8 | 0.000000 | 1.8 | 11.193277 |
Exercise 2.2¶
Using the meteorite data from the Meteorite_Landings.csv file, update the year column to only contain the year, convert it to a numeric data type, and create a new column indicating whether the meteorite was observed falling before 1970. Set the index to the id column and extract all the rows with IDs between 10,036 and 10,040 (inclusive) with loc[].¶
Hint 1: Use year.str.slice() to grab a substring.¶
Hint 2: Make sure to sort the index before using loc[] to select the range.¶
Bonus: There's a data entry error in the year column. Can you find it? (Don't spend too much time on this.)¶
Solution¶
import pandas as pd
meteorites = pd.read_csv('../data/Meteorite_Landings.csv').assign(
year=lambda x: pd.to_numeric(x.year.str.slice(6, 10)),
pre_1970=lambda x: (x.fall == 'Fell') & (x.year < 1970)
).set_index('id')
meteorites.sort_index().loc[10_036:10_040]
| name | nametype | recclass | mass (g) | fall | year | reclat | reclong | GeoLocation | pre_1970 | |
|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||
| 10036 | Enigma | Valid | H4 | 94.0 | Found | 1967.0 | 31.33333 | -82.31667 | (31.33333, -82.31667) | False |
| 10037 | Enon | Valid | Iron, ungrouped | 763.0 | Found | 1883.0 | 39.86667 | -83.95000 | (39.86667, -83.95) | False |
| 10038 | Enshi | Valid | H5 | 8000.0 | Fell | 1974.0 | 30.30000 | 109.50000 | (30.3, 109.5) | False |
| 10039 | Ensisheim | Valid | LL6 | 127000.0 | Fell | 1491.0 | 47.86667 | 7.35000 | (47.86667, 7.35) | True |
Note: The pd.to_datetime() function is another option here; however, it will only be able to convert dates within the supported bounds (between pd.Timestamp.min and pd.Timestamp.max), which will cause some entries that do have a year to be marked as not having one. More information can be found in the pandas documentation here. For reference, this is how the conversion can be done:
pd.to_datetime(
meteorites.year,
errors='coerce', # anything that can't be converted will be NaT (null)
format='%m/%d/%Y %I:%M:%S %p' # the format the datetimes are currently in
)
Bonus: There's a data entry error in the year column. Can you find it?¶
meteorites.year.describe()
count 45425.000000 mean 1991.828817 std 25.052766 min 860.000000 25% 1987.000000 50% 1998.000000 75% 2003.000000 max 2101.000000 Name: year, dtype: float64
There's a meteorite that was reportedly found in the future:
meteorites.query(f'year > {pd.Timestamp("today").year}')
| name | nametype | recclass | mass (g) | fall | year | reclat | reclong | GeoLocation | pre_1970 | |
|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||
| 57150 | Northwest Africa 7701 | Valid | CK6 | 55.0 | Found | 2101.0 | 0.0 | 0.0 | (0.0, 0.0) | False |
Learning Path¶
- Data cleaning
- Working with the index
- Reshaping data
- Aggregations and grouping
- Time series
Reshaping data¶
The taxi dataset we have be working with is in a format conducive to an analysis. This isn't always the case. Let's now take a look at the TSA traveler throughput data, which compares 2021 throughput to the same day in 2020 and 2019:
tsa = pd.read_csv('../data/tsa_passenger_throughput.csv', parse_dates=['Date'])
tsa.head()
| Date | 2021 Traveler Throughput | 2020 Traveler Throughput | 2019 Traveler Throughput | |
|---|---|---|---|---|
| 0 | 2021-05-14 | 1716561.0 | 250467 | 2664549 |
| 1 | 2021-05-13 | 1743515.0 | 234928 | 2611324 |
| 2 | 2021-05-12 | 1424664.0 | 176667 | 2343675 |
| 3 | 2021-05-11 | 1315493.0 | 163205 | 2191387 |
| 4 | 2021-05-10 | 1657722.0 | 215645 | 2512315 |
Source: TSA.gov (collected on May 16, 2021)
First, we will lowercase the column names and take the first word (e.g., 2021 for 2021 Traveler Throughput) to make this easier to work with:
tsa = tsa.rename(columns=lambda x: x.lower().split()[0])
tsa.head()
| date | 2021 | 2020 | 2019 | |
|---|---|---|---|---|
| 0 | 2021-05-14 | 1716561.0 | 250467 | 2664549 |
| 1 | 2021-05-13 | 1743515.0 | 234928 | 2611324 |
| 2 | 2021-05-12 | 1424664.0 | 176667 | 2343675 |
| 3 | 2021-05-11 | 1315493.0 | 163205 | 2191387 |
| 4 | 2021-05-10 | 1657722.0 | 215645 | 2512315 |
Now, we can work on reshaping it into two columns: the date and the traveler throughput from 2019 through 2021.
Starting with the long-format data below, we want to melt it into wide-format data so that we can look at the evolution of the throughput over time:
from utils import highlight_long_format
colors = {'2021': 'pink', '2020': 'skyblue', '2019': 'lightgreen'}
highlight_long_format(tsa.head(2), colors)
| date | 2021 | 2020 | 2019 | |
|---|---|---|---|---|
| 0 | 2021-05-14 00:00:00 | 1716561.000000 | 250467 | 2664549 |
| 1 | 2021-05-13 00:00:00 | 1743515.000000 | 234928 | 2611324 |
Note that the two rows above contain the same data as the six rows below:
from utils import highlight_wide_format
highlight_wide_format(tsa.head(2), colors)
| date | year | travelers | |
|---|---|---|---|
| 0 | 2021-05-14 00:00:00 | 2021 | 1716561.000000 |
| 1 | 2021-05-13 00:00:00 | 2021 | 1743515.000000 |
| 2 | 2020-05-14 00:00:00 | 2020 | 250467.000000 |
| 3 | 2020-05-13 00:00:00 | 2020 | 234928.000000 |
| 4 | 2019-05-14 00:00:00 | 2019 | 2664549.000000 |
| 5 | 2019-05-13 00:00:00 | 2019 | 2611324.000000 |
Let's work on making this transformation.
Melting¶
Melting helps convert our data into long format. Now, we have all the traveler throughput numbers in a single column:
tsa_melted = tsa.melt(
id_vars='date', # column that uniquely identifies a row (can be multiple)
var_name='year', # name for the new column created by melting
value_name='travelers' # name for new column containing values from melted columns
)
tsa_melted.sample(5, random_state=1) # show some random entries
| date | year | travelers | |
|---|---|---|---|
| 974 | 2020-09-12 | 2019 | 1879822.0 |
| 435 | 2021-03-05 | 2020 | 2198517.0 |
| 1029 | 2020-07-19 | 2019 | 2727355.0 |
| 680 | 2020-07-03 | 2020 | 718988.0 |
| 867 | 2020-12-28 | 2019 | 2500396.0 |
To convert this into a time series of traveler throughput, we need to replace the year in the date column with the one in the year column. Otherwise, we are marking prior years' numbers with the wrong year.
tsa_melted = tsa_melted.assign(
date=lambda x: pd.to_datetime(x.year + x.date.dt.strftime('-%m-%d'))
)
tsa_melted.sample(5, random_state=1)
| date | year | travelers | |
|---|---|---|---|
| 974 | 2019-09-12 | 2019 | 1879822.0 |
| 435 | 2020-03-05 | 2020 | 2198517.0 |
| 1029 | 2019-07-19 | 2019 | 2727355.0 |
| 680 | 2020-07-03 | 2020 | 718988.0 |
| 867 | 2019-12-28 | 2019 | 2500396.0 |
This leaves us with some null values (the dates that aren't present in the dataset):
tsa_melted.sort_values('date').tail(3)
| date | year | travelers | |
|---|---|---|---|
| 136 | 2021-12-29 | 2021 | NaN |
| 135 | 2021-12-30 | 2021 | NaN |
| 134 | 2021-12-31 | 2021 | NaN |
These can be dropped with the dropna() method:
tsa_melted = tsa_melted.dropna()
tsa_melted.sort_values('date').tail(3)
| date | year | travelers | |
|---|---|---|---|
| 2 | 2021-05-12 | 2021 | 1424664.0 |
| 1 | 2021-05-13 | 2021 | 1743515.0 |
| 0 | 2021-05-14 | 2021 | 1716561.0 |
Pivoting¶
Using the melted data, we can pivot the data to compare TSA traveler throughput on specific days across years:
tsa_pivoted = tsa_melted\
.query('date.dt.month == 3 and date.dt.day <= 10')\
.assign(day_in_march=lambda x: x.date.dt.day)\
.pivot(index='year', columns='day_in_march', values='travelers')
tsa_pivoted
| day_in_march | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| year | ||||||||||
| 2019 | 2257920.0 | 1979558.0 | 2143619.0 | 2402692.0 | 2543689.0 | 2156262.0 | 2485430.0 | 2378673.0 | 2122898.0 | 2187298.0 |
| 2020 | 2089641.0 | 1736393.0 | 1877401.0 | 2130015.0 | 2198517.0 | 1844811.0 | 2119867.0 | 1909363.0 | 1617220.0 | 1702686.0 |
| 2021 | 1049692.0 | 744812.0 | 826924.0 | 1107534.0 | 1168734.0 | 992406.0 | 1278557.0 | 1119303.0 | 825745.0 | 974221.0 |
Important: We aren't covering the unstack() and stack() methods, which are additional ways to pivot and melt, respectively. These come in handy when we have a multi-level index (e.g., if we ran set_index() with more than one column). More information can be found here.
Transposing¶
The T attribute provides a quick way to flip rows and columns.
tsa_pivoted.T
| year | 2019 | 2020 | 2021 |
|---|---|---|---|
| day_in_march | |||
| 1 | 2257920.0 | 2089641.0 | 1049692.0 |
| 2 | 1979558.0 | 1736393.0 | 744812.0 |
| 3 | 2143619.0 | 1877401.0 | 826924.0 |
| 4 | 2402692.0 | 2130015.0 | 1107534.0 |
| 5 | 2543689.0 | 2198517.0 | 1168734.0 |
| 6 | 2156262.0 | 1844811.0 | 992406.0 |
| 7 | 2485430.0 | 2119867.0 | 1278557.0 |
| 8 | 2378673.0 | 1909363.0 | 1119303.0 |
| 9 | 2122898.0 | 1617220.0 | 825745.0 |
| 10 | 2187298.0 | 1702686.0 | 974221.0 |
Merging¶
We typically observe changes in air travel around the holidays, so adding information about the dates in the TSA dataset provides more context. The holidays.csv file contains a few major holidays in the United States:
holidays = pd.read_csv('../data/holidays.csv', parse_dates=True, index_col='date')
holidays.loc['2019']
| holiday | |
|---|---|
| date | |
| 2019-01-01 | New Year's Day |
| 2019-05-27 | Memorial Day |
| 2019-07-04 | July 4th |
| 2019-09-02 | Labor Day |
| 2019-11-28 | Thanksgiving |
| 2019-12-24 | Christmas Eve |
| 2019-12-25 | Christmas Day |
| 2019-12-31 | New Year's Eve |
Merging the holidays with the TSA traveler throughput data will provide more context for our analysis:
tsa_melted_holidays = tsa_melted\
.merge(holidays, left_on='date', right_index=True, how='left')\
.sort_values('date')
tsa_melted_holidays.head()
| date | year | travelers | holiday | |
|---|---|---|---|---|
| 863 | 2019-01-01 | 2019 | 2126398.0 | New Year's Day |
| 862 | 2019-01-02 | 2019 | 2345103.0 | NaN |
| 861 | 2019-01-03 | 2019 | 2202111.0 | NaN |
| 860 | 2019-01-04 | 2019 | 2150571.0 | NaN |
| 859 | 2019-01-05 | 2019 | 1975947.0 | NaN |
Tip: There are many parameters for this method, so be sure to check out the documentation. To append rows, take a look at the pd.concat() function.
We can take this a step further by marking a few days before and after each holiday as part of the holiday. This would make it easier to compare holiday travel across years and look for any uptick in travel around the holidays:
tsa_melted_holiday_travel = tsa_melted_holidays.assign(
holiday=lambda x:
x.holiday\
.ffill(limit=1)\
.bfill(limit=2)
)
Tip: Check out the fillna() method documentation for additional functionality for replacing NA/NaN values.
Notice that we now have values for the day after each holiday and the two days prior. Thanksgiving in 2019 was on November 28th, so the 26th, 27th, and 29th were filled. Since we are only replacing null values, we don't override Christmas Day with the forward fill of Christmas Eve:
tsa_melted_holiday_travel.query(
'year == "2019" and '
'(holiday == "Thanksgiving" or holiday.str.contains("Christmas"))'
)
| date | year | travelers | holiday | |
|---|---|---|---|---|
| 899 | 2019-11-26 | 2019 | 1591158.0 | Thanksgiving |
| 898 | 2019-11-27 | 2019 | 1968137.0 | Thanksgiving |
| 897 | 2019-11-28 | 2019 | 2648268.0 | Thanksgiving |
| 896 | 2019-11-29 | 2019 | 2882915.0 | Thanksgiving |
| 873 | 2019-12-22 | 2019 | 1981433.0 | Christmas Eve |
| 872 | 2019-12-23 | 2019 | 1937235.0 | Christmas Eve |
| 871 | 2019-12-24 | 2019 | 2552194.0 | Christmas Eve |
| 870 | 2019-12-25 | 2019 | 2582580.0 | Christmas Day |
| 869 | 2019-12-26 | 2019 | 2470786.0 | Christmas Day |
Learning Path¶
- Data cleaning
- Working with the index
- Reshaping data
- Aggregations and grouping
- Time series
Aggregations and grouping¶
After reshaping and cleaning our data, we can perform aggregations to summarize it in a variety of ways. In this section, we will explore using pivot tables, crosstabs, and group by operations to aggregate the data.
Pivot tables¶
We can build a pivot table to compare holiday travel across the years in our dataset:
tsa_melted_holiday_travel.pivot_table(
index='year', columns='holiday', sort=False,
values='travelers', aggfunc='sum'
)
| holiday | New Year's Day | Memorial Day | July 4th | Labor Day | Thanksgiving | Christmas Eve | Christmas Day | New Year's Eve |
|---|---|---|---|---|---|---|---|---|
| year | ||||||||
| 2019 | 4471501.0 | 9720691.0 | 9414228.0 | 8314811.0 | 9090478.0 | 6470862.0 | 5053366.0 | 6535464.0 |
| 2020 | 4490388.0 | 1126253.0 | 2682541.0 | 2993653.0 | 3364358.0 | 3029810.0 | 1745242.0 | 3057449.0 |
| 2021 | 1998871.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
We can use the pct_change() method on this result to see which holiday travel periods saw the biggest change in travel:
tsa_melted_holiday_travel.pivot_table(
index='year', columns='holiday', sort=False,
values='travelers', aggfunc='sum'
).pct_change(fill_method=None)
| holiday | New Year's Day | Memorial Day | July 4th | Labor Day | Thanksgiving | Christmas Eve | Christmas Day | New Year's Eve |
|---|---|---|---|---|---|---|---|---|
| year | ||||||||
| 2019 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 2020 | 0.004224 | -0.884139 | -0.715055 | -0.639961 | -0.629903 | -0.531776 | -0.654638 | -0.532176 |
| 2021 | -0.554856 | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
Let's make one last pivot table with column and row subtotals, along with some formatting improvements. First, we set a display option for all floats:
pd.set_option('display.float_format', '{:,.0f}'.format)
Next, we group together Christmas Eve and Christmas Day, likewise for New Year's Eve and New Year's Day (handling for the change in year), and create the pivot table:
import numpy as np
pivot_table = tsa_melted_holiday_travel.assign(
year=lambda x: np.where(
x.holiday == "New Year's Day", pd.to_numeric(x.year) - 1, x.year
).astype(str),
holiday=lambda x: np.where(
x.holiday.str.contains('Christmas|New Year', regex=True),
x.holiday.str.replace('Day|Eve', '', regex=True).str.strip(),
x.holiday
)
).pivot_table(
index='year', columns='holiday', sort=False,
values='travelers', aggfunc='sum',
margins=True, margins_name='Total'
)
# reorder columns by order in the year
pivot_table.insert(5, "New Year's", pivot_table.pop("New Year's"))
pivot_table
| holiday | Memorial Day | July 4th | Labor Day | Thanksgiving | Christmas | New Year's | Total |
|---|---|---|---|---|---|---|---|
| year | |||||||
| 2018 | NaN | NaN | NaN | NaN | NaN | 4,471,501 | 4,471,501 |
| 2019 | 9,720,691 | 9,414,228 | 8,314,811 | 9,090,478 | 11,524,228 | 11,025,852 | 59,090,288 |
| 2020 | 1,126,253 | 2,682,541 | 2,993,653 | 3,364,358 | 4,775,052 | 5,056,320 | 19,998,177 |
| Total | 10,846,944 | 12,096,769 | 11,308,464 | 12,454,836 | 16,299,280 | 20,553,673 | 83,559,966 |
Before moving on, let's reset the display option:
pd.reset_option('display.float_format')
Tip: Read more about options in the documentation here.
Exercise 2.3¶
Using the meteorite data from the Meteorite_Landings.csv file, create a pivot table that shows both the number of meteorites and the 95th percentile of meteorite mass for those that were found versus observed falling per year from 2005 through 2009 (inclusive). Hint: Be sure to convert the year column to a number as we did in the previous exercise.¶
Solution¶
import pandas as pd
meteorites = pd.read_csv('../data/Meteorite_Landings.csv').assign(
year=lambda x: pd.to_numeric(x.year.str.slice(6, 10))
)
meteorites.query('year.between(2005, 2009)').pivot_table(
index='year', columns='fall', values='mass (g)',
aggfunc=['count', lambda x: x.quantile(0.95)]
).rename(columns={'<lambda>': '95th percentile'})
| count | 95th percentile | |||
|---|---|---|---|---|
| fall | Fell | Found | Fell | Found |
| year | ||||
| 2005.0 | NaN | 874.0 | NaN | 4500.00 |
| 2006.0 | 5.0 | 2450.0 | 25008.0 | 1600.50 |
| 2007.0 | 8.0 | 1181.0 | 89675.0 | 1126.90 |
| 2008.0 | 9.0 | 948.0 | 106000.0 | 2274.80 |
| 2009.0 | 5.0 | 1492.0 | 8333.4 | 1397.25 |
Crosstabs¶
The pd.crosstab() function provides an easy way to create a frequency table. Here, we count the number of low-, medium-, and high-volume travel days per year, using the pd.cut() function to create three travel volume bins of equal width:
pd.crosstab(
index=pd.cut(
tsa_melted_holiday_travel.travelers,
bins=3, labels=['low', 'medium', 'high']
),
columns=tsa_melted_holiday_travel.year,
rownames=['travel_volume']
)
| year | 2019 | 2020 | 2021 |
|---|---|---|---|
| travel_volume | |||
| low | 0 | 277 | 54 |
| medium | 42 | 44 | 80 |
| high | 323 | 44 | 0 |
Tip: The pd.cut() function can also be used to specify custom bin ranges. For equal-sized bins based on quantiles, use the pd.qcut() function instead.
Note that the pd.crosstab() function supports other aggregations provided you pass in the data to aggregate as values and specify the aggregation with aggfunc. You can also add subtotals and normalize the data. See the documentation for more information.
Group by operations¶
Rather than perform aggregations, like mean() or describe(), on the full dataset at once, we can perform these calculations per group by first calling groupby():
tsa_melted_holiday_travel.groupby('year').describe(include=np.number)
| travelers | ||||||||
|---|---|---|---|---|---|---|---|---|
| count | mean | std | min | 25% | 50% | 75% | max | |
| year | ||||||||
| 2019 | 365.0 | 2.309482e+06 | 285061.490784 | 1534386.0 | 2091116.0 | 2358007.0 | 2538384.00 | 2882915.0 |
| 2020 | 365.0 | 8.818674e+05 | 639775.194297 | 87534.0 | 507129.0 | 718310.0 | 983745.00 | 2507588.0 |
| 2021 | 134.0 | 1.112632e+06 | 338040.673782 | 468933.0 | 807156.0 | 1117391.0 | 1409377.75 | 1743515.0 |
Groups can also be used to perform separate calculations per subset of the data. For example, we can find the highest-volume travel day per year using rank():
tsa_melted_holiday_travel.assign(
travel_volume_rank=lambda x: x.groupby('year').travelers.rank(ascending=False)
).sort_values(['travel_volume_rank', 'year']).head(3)
| date | year | travelers | holiday | travel_volume_rank | |
|---|---|---|---|---|---|
| 896 | 2019-11-29 | 2019 | 2882915.0 | Thanksgiving | 1.0 |
| 456 | 2020-02-12 | 2020 | 2507588.0 | NaN | 1.0 |
| 1 | 2021-05-13 | 2021 | 1743515.0 | NaN | 1.0 |
The previous two examples called a single method on the grouped data, but using the agg() method we can specify any number of them:
tsa_melted_holiday_travel.assign(
holiday_travelers=lambda x: np.where(~x.holiday.isna(), x.travelers, np.nan),
non_holiday_travelers=lambda x: np.where(x.holiday.isna(), x.travelers, np.nan),
year=lambda x: pd.to_numeric(x.year)
).select_dtypes(include='number').groupby('year').agg(['mean', 'std'])
| travelers | holiday_travelers | non_holiday_travelers | ||||
|---|---|---|---|---|---|---|
| mean | std | mean | std | mean | std | |
| year | ||||||
| 2019 | 2.309482e+06 | 285061.490784 | 2.271977e+06 | 303021.675751 | 2.312359e+06 | 283906.226598 |
| 2020 | 8.818674e+05 | 639775.194297 | 8.649882e+05 | 489938.240989 | 8.831619e+05 | 650399.772930 |
| 2021 | 1.112632e+06 | 338040.673782 | 9.994355e+05 | 273573.249680 | 1.114347e+06 | 339479.298658 |
Tip: The select_dtypes() method makes it possible to select columns by their data type. We can specify the data types to exclude and/or include.
In addition, we can specify which aggregations to perform on each column:
tsa_melted_holiday_travel.assign(
holiday_travelers=lambda x: np.where(~x.holiday.isna(), x.travelers, np.nan),
non_holiday_travelers=lambda x: np.where(x.holiday.isna(), x.travelers, np.nan)
).groupby('year').agg({
'holiday_travelers': ['mean', 'std'],
'holiday': ['nunique', 'count']
})
| holiday_travelers | holiday | |||
|---|---|---|---|---|
| mean | std | nunique | count | |
| year | ||||
| 2019 | 2.271977e+06 | 303021.675751 | 8 | 26 |
| 2020 | 8.649882e+05 | 489938.240989 | 8 | 26 |
| 2021 | 9.994355e+05 | 273573.249680 | 1 | 2 |
We are only scratching the surface; some additional functionalities to be aware of include the following:
- We can group by multiple columns – this creates a hierarchical index.
- Groups can be excluded from calculations with the
filter()method. - We can group on content in the index using the
levelornameparameters e.g.,groupby(level=0)orgroupby(name='year'). - We can group by date ranges if we use a
pd.Grouper()object.
Be sure to check out the documentation for more details.
Solution¶
import pandas as pd
meteorites = pd.read_csv('../data/Meteorite_Landings.csv')
meteorites.groupby('fall')['mass (g)'].describe()
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| fall | ||||||||
| Fell | 1075.0 | 47070.715023 | 717067.125826 | 0.1 | 686.00 | 2800.0 | 10450.0 | 23000000.0 |
| Found | 44510.0 | 12461.922983 | 571105.752311 | 0.0 | 6.94 | 30.5 | 178.0 | 60000000.0 |
Learning Path¶
- Data cleaning
- Working with the index
- Reshaping data
- Aggregations and grouping
- Time series
Time series¶
When working with time series data, pandas provides us with additional functionality to not just compare the observations in our dataset, but to use their relationship in time to analyze the data. In this section, we will see a few such operations for selecting date/time ranges, calculating changes over time, performing window calculations, and resampling the data to different date/time intervals.
Selecting based on date and time¶
Let's switch back to the taxis dataset, which has timestamps of pickups and dropoffs. First, we will set the dropoff column as the index and sort the data:
taxis = taxis.set_index('dropoff').sort_index()
We saw earlier that we can slice on the datetimes:
taxis['2019-10-24 12':'2019-10-24 13']
| pickup | passenger_count | trip_distance | payment_type | fare_amount | extra | mta_tax | tip_amount | tolls_amount | improvement_surcharge | total_amount | congestion_surcharge | elapsed_time | cost_before_tip | tip_pct | fees | avg_speed | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dropoff | |||||||||||||||||
| 2019-10-24 12:30:08 | 2019-10-23 13:25:42 | 4 | 0.76 | 2 | 5.0 | 1.0 | 0.5 | 0.00 | 0.0 | 0.3 | 9.30 | 2.5 | 0 days 23:04:26 | 9.3 | 0.0 | 4.3 | 0.032938 |
| 2019-10-24 12:42:01 | 2019-10-23 13:34:03 | 2 | 1.58 | 1 | 7.5 | 1.0 | 0.5 | 2.36 | 0.0 | 0.3 | 14.16 | 2.5 | 0 days 23:07:58 | 11.8 | 0.2 | 4.3 | 0.068301 |
We can also represent this range with shorthand. Note that we must use loc[] here:
taxis.loc['2019-10-24 12']
| pickup | passenger_count | trip_distance | payment_type | fare_amount | extra | mta_tax | tip_amount | tolls_amount | improvement_surcharge | total_amount | congestion_surcharge | elapsed_time | cost_before_tip | tip_pct | fees | avg_speed | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dropoff | |||||||||||||||||
| 2019-10-24 12:30:08 | 2019-10-23 13:25:42 | 4 | 0.76 | 2 | 5.0 | 1.0 | 0.5 | 0.00 | 0.0 | 0.3 | 9.30 | 2.5 | 0 days 23:04:26 | 9.3 | 0.0 | 4.3 | 0.032938 |
| 2019-10-24 12:42:01 | 2019-10-23 13:34:03 | 2 | 1.58 | 1 | 7.5 | 1.0 | 0.5 | 2.36 | 0.0 | 0.3 | 14.16 | 2.5 | 0 days 23:07:58 | 11.8 | 0.2 | 4.3 | 0.068301 |
However, if we want to look at this time range across days, we need another strategy.
We can pull out the dropoffs that happened between a certain time range on any day with the between_time() method:
taxis.between_time('12:00', '13:00')
| pickup | passenger_count | trip_distance | payment_type | fare_amount | extra | mta_tax | tip_amount | tolls_amount | improvement_surcharge | total_amount | congestion_surcharge | elapsed_time | cost_before_tip | tip_pct | fees | avg_speed | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dropoff | |||||||||||||||||
| 2019-10-23 12:53:49 | 2019-10-23 12:35:27 | 5 | 2.49 | 1 | 13.5 | 1.0 | 0.5 | 2.20 | 0.0 | 0.3 | 20.00 | 2.5 | 0 days 00:18:22 | 17.8 | 0.123596 | 4.3 | 8.134301 |
| 2019-10-24 12:30:08 | 2019-10-23 13:25:42 | 4 | 0.76 | 2 | 5.0 | 1.0 | 0.5 | 0.00 | 0.0 | 0.3 | 9.30 | 2.5 | 0 days 23:04:26 | 9.3 | 0.000000 | 4.3 | 0.032938 |
| 2019-10-24 12:42:01 | 2019-10-23 13:34:03 | 2 | 1.58 | 1 | 7.5 | 1.0 | 0.5 | 2.36 | 0.0 | 0.3 | 14.16 | 2.5 | 0 days 23:07:58 | 11.8 | 0.200000 | 4.3 | 0.068301 |
Tip: The at_time() method can be used to extract all entries at a given time (e.g., 12:35:27).
For the rest of this section, we will be working with the TSA traveler throughput data. Let's start by setting the index to the date column:
tsa_melted_holiday_travel = tsa_melted_holiday_travel.set_index('date')
Calculating change over time¶
tsa_melted_holiday_travel.loc['2020'].assign(
one_day_change=lambda x: x.travelers.diff(),
seven_day_change=lambda x: x.travelers.diff(7),
).head(10)
| year | travelers | holiday | one_day_change | seven_day_change | |
|---|---|---|---|---|---|
| date | |||||
| 2020-01-01 | 2020 | 2311732.0 | New Year's Day | NaN | NaN |
| 2020-01-02 | 2020 | 2178656.0 | New Year's Day | -133076.0 | NaN |
| 2020-01-03 | 2020 | 2422272.0 | NaN | 243616.0 | NaN |
| 2020-01-04 | 2020 | 2210542.0 | NaN | -211730.0 | NaN |
| 2020-01-05 | 2020 | 1806480.0 | NaN | -404062.0 | NaN |
| 2020-01-06 | 2020 | 1815040.0 | NaN | 8560.0 | NaN |
| 2020-01-07 | 2020 | 2034472.0 | NaN | 219432.0 | NaN |
| 2020-01-08 | 2020 | 2072543.0 | NaN | 38071.0 | -239189.0 |
| 2020-01-09 | 2020 | 1687974.0 | NaN | -384569.0 | -490682.0 |
| 2020-01-10 | 2020 | 2183734.0 | NaN | 495760.0 | -238538.0 |
Tip: To perform operations other than subtraction, take a look at the shift() method. It also makes it possible to perform operations across columns.
tsa_melted_holiday_travel['2019':'2021-Q1'].select_dtypes(include='number')\
.resample('QE').agg(['sum', 'mean', 'std'])
| travelers | |||
|---|---|---|---|
| sum | mean | std | |
| date | |||
| 2019-03-31 | 189281658.0 | 2.103130e+06 | 282239.618354 |
| 2019-06-30 | 221756667.0 | 2.436886e+06 | 212600.697665 |
| 2019-09-30 | 220819236.0 | 2.400209e+06 | 260140.242892 |
| 2019-12-31 | 211103512.0 | 2.294603e+06 | 260510.040655 |
| 2020-03-31 | 155354148.0 | 1.726157e+06 | 685094.277420 |
| 2020-06-30 | 25049083.0 | 2.752646e+05 | 170127.402046 |
| 2020-09-30 | 63937115.0 | 6.949686e+05 | 103864.705739 |
| 2020-12-31 | 77541248.0 | 8.428397e+05 | 170245.484185 |
| 2021-03-31 | 86094635.0 | 9.566071e+05 | 280399.809061 |
Window calculations¶
Window calculations are similar to group by calculations except the group over which the calculation is performed isn't static – it can move or expand. Pandas provides functionality for constructing a variety of windows, including moving/rolling windows, expanding windows (e.g., cumulative sum or mean up to the current date in a time series), and exponentially weighted moving windows (to weight closer observations more than further ones). We will only look at rolling and expanding calculations here.
Performing a window calculation is very similar to a group by calculation – we first define the window, and then we specify the aggregation:
tsa_melted_holiday_travel.loc['2020'].assign(
**{
'7D MA': lambda x: x.rolling('7D').travelers.mean(),
'YTD mean': lambda x: x.expanding().travelers.mean()
}
).head(10)
| year | travelers | holiday | 7D MA | YTD mean | |
|---|---|---|---|---|---|
| date | |||||
| 2020-01-01 | 2020 | 2311732.0 | New Year's Day | 2.311732e+06 | 2.311732e+06 |
| 2020-01-02 | 2020 | 2178656.0 | New Year's Day | 2.245194e+06 | 2.245194e+06 |
| 2020-01-03 | 2020 | 2422272.0 | NaN | 2.304220e+06 | 2.304220e+06 |
| 2020-01-04 | 2020 | 2210542.0 | NaN | 2.280800e+06 | 2.280800e+06 |
| 2020-01-05 | 2020 | 1806480.0 | NaN | 2.185936e+06 | 2.185936e+06 |
| 2020-01-06 | 2020 | 1815040.0 | NaN | 2.124120e+06 | 2.124120e+06 |
| 2020-01-07 | 2020 | 2034472.0 | NaN | 2.111313e+06 | 2.111313e+06 |
| 2020-01-08 | 2020 | 2072543.0 | NaN | 2.077144e+06 | 2.106467e+06 |
| 2020-01-09 | 2020 | 1687974.0 | NaN | 2.007046e+06 | 2.059968e+06 |
| 2020-01-10 | 2020 | 2183734.0 | NaN | 1.972969e+06 | 2.072344e+06 |
To understand what's happening, it's best to visualize the original data and the result, so here's a sneak peek of plotting with pandas. First, some setup to embed SVG-format plots in the notebook:
import matplotlib_inline
from utils import mpl_svg_config
matplotlib_inline.backend_inline.set_matplotlib_formats(
'svg', # output images using SVG format
**mpl_svg_config('section-2') # optional: configure metadata
)
Tip: For most use cases, only the first argument is necessary – we will discuss the second argument in more detail in the next section.
Now, we call the plot() method to visualize the data:
_ = tsa_melted_holiday_travel.loc['2020'].assign(
**{
'7D MA': lambda x: x.rolling('7D').travelers.mean(),
'YTD mean': lambda x: x.expanding().travelers.mean()
}
).plot(title='2020 TSA Traveler Throughput', ylabel='travelers', alpha=0.8)
Other types of windows:
- exponentially weighted moving: use the
ewm()method - custom: create a subclass of
pandas.api.indexers.BaseIndexeror use a pre-built one inpandas.api.indexers
Solution¶
import pandas as pd
taxis = pd.read_csv(
'../data/2019_Yellow_Taxi_Trip_Data.csv',
parse_dates=True, index_col='tpep_dropoff_datetime'
)
taxis.resample('1h')[[
'trip_distance', 'fare_amount', 'tolls_amount', 'tip_amount'
]].sum().nlargest(5, 'tip_amount')
| trip_distance | fare_amount | tolls_amount | tip_amount | |
|---|---|---|---|---|
| tpep_dropoff_datetime | ||||
| 2019-10-23 16:00:00 | 10676.95 | 67797.76 | 699.04 | 12228.64 |
| 2019-10-23 17:00:00 | 16052.83 | 70131.91 | 4044.04 | 12044.03 |
| 2019-10-23 18:00:00 | 3104.56 | 11565.56 | 1454.67 | 1907.64 |
| 2019-10-23 15:00:00 | 14.34 | 213.50 | 0.00 | 51.75 |
| 2019-10-23 19:00:00 | 98.59 | 268.00 | 24.48 | 25.74 |
Section 2 Complete 🎉¶

Section 3: Data Visualization¶
The human brain excels at finding patterns in visual representations of the data; so in this section, we will learn how to visualize data using pandas along with the Matplotlib and Seaborn libraries for additional features. We will create a variety of visualizations that will help us better understand our data.
Learning Path¶
- Why is data visualization necessary?
- Plotting with pandas
- Plotting with Seaborn
- Customizing plots with Matplotlib
Why is data visualization necessary?¶
So far, we have focused a lot on summarizing the data using statistics. However, summary statistics are not enough to understand the distribution – there are many possible distributions for a given set of summary statistics. Data visualization is necessary to truly understand the distribution:
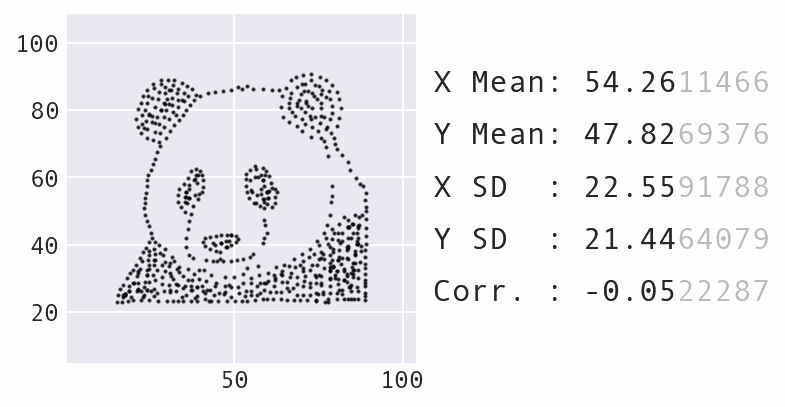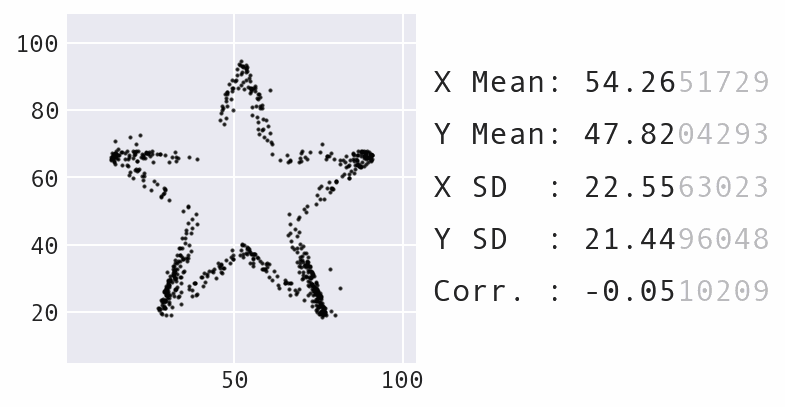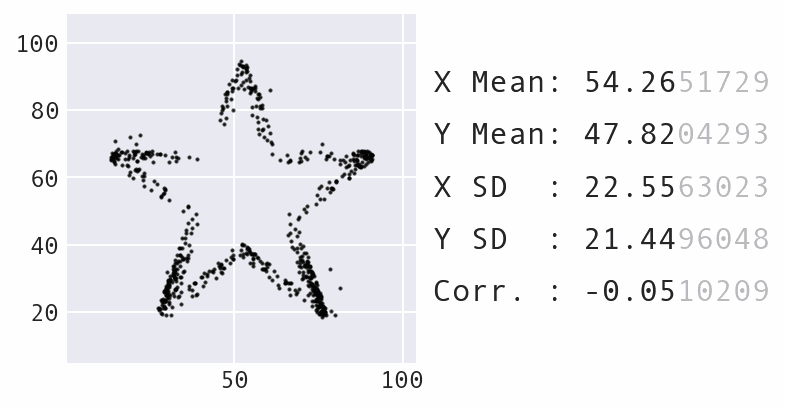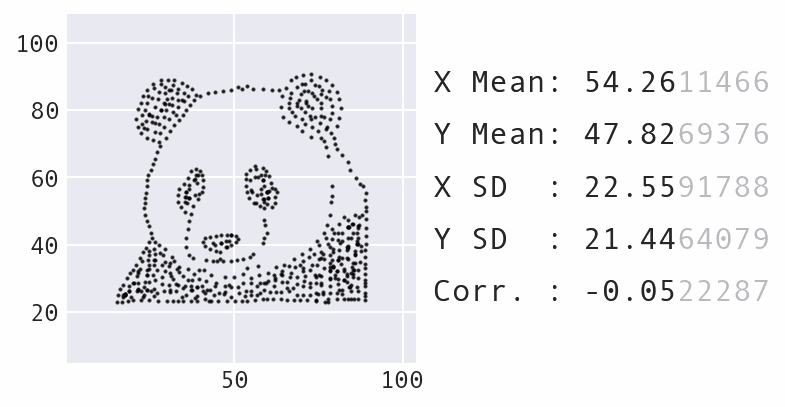
Learning Path¶
- Why is data visualization necessary?
- Plotting with pandas
- Plotting with Seaborn
- Customizing plots with Matplotlib
Plotting with pandas¶
We can create a variety of visualizations using the plot() method. In this section, we will take a brief tour of some of this functionality, which under the hood uses Matplotlib.
Once again, we will be working with the TSA traveler throughput data that we cleaned up in the previous section:
import pandas as pd
tsa_melted_holiday_travel = pd.read_csv(
'../data/tsa_melted_holiday_travel.csv',
parse_dates=True, index_col='date'
)
tsa_melted_holiday_travel.head()
| year | travelers | holiday | |
|---|---|---|---|
| date | |||
| 2019-01-01 | 2019 | 2126398.0 | New Year's Day |
| 2019-01-02 | 2019 | 2345103.0 | New Year's Day |
| 2019-01-03 | 2019 | 2202111.0 | NaN |
| 2019-01-04 | 2019 | 2150571.0 | NaN |
| 2019-01-05 | 2019 | 1975947.0 | NaN |
To embed SVG-format plots in the notebook, we will configure the Matplotlib plotting backend to generate SVG output (first argument) with custom metadata (second argument):
import matplotlib_inline
from utils import mpl_svg_config
matplotlib_inline.backend_inline.set_matplotlib_formats(
'svg', # output images using SVG format
**mpl_svg_config('section-3') # optional: configure metadata
)
Note: The second argument is optional and is used here to make the SVG output reproducible by setting the hashsalt along with some metadata, which will be used by Matplotlib when generating any SVG output (see the utils.py file for more details). Without this argument, different runs of the same plotting code will generate plots that are visually identical, but differ at the HTML level due to different IDs, metadata, etc.
Line plots¶
Let's continue with the example of rolling and expanding calculations:
plot_data = tsa_melted_holiday_travel.drop(columns='year').loc['2020'].assign(
**{
'7D MA': lambda x: x.travelers.rolling('7D').mean(),
'YTD mean': lambda x: x.travelers.expanding().mean()
}
)
plot_data.head()
| travelers | holiday | 7D MA | YTD mean | |
|---|---|---|---|---|
| date | ||||
| 2020-01-01 | 2311732.0 | New Year's Day | 2311732.0 | 2311732.0 |
| 2020-01-02 | 2178656.0 | New Year's Day | 2245194.0 | 2245194.0 |
| 2020-01-03 | 2422272.0 | NaN | 2304220.0 | 2304220.0 |
| 2020-01-04 | 2210542.0 | NaN | 2280800.5 | 2280800.5 |
| 2020-01-05 | 1806480.0 | NaN | 2185936.4 | 2185936.4 |
The plot() method will generate line plots for all numeric columns by default:
plot_data.plot(
title='2020 TSA Traveler Throughput', ylabel='travelers', alpha=0.8
)
<Axes: title={'center': '2020 TSA Traveler Throughput'}, xlabel='date', ylabel='travelers'>
The plot() method returns an Axes object that can be modified further (e.g., to add reference lines, annotations, labels, etc.). Let's walk through an example.
Bar plots¶
For our next example, we will plot vertical bars to compare monthly TSA traveler throughput across years. Let's start by creating a pivot table with the information we need:
plot_data = tsa_melted_holiday_travel['2019':'2021-04']\
.assign(month=lambda x: x.index.month)\
.pivot_table(index='month', columns='year', values='travelers', aggfunc='sum')
plot_data.head()
| year | 2019 | 2020 | 2021 |
|---|---|---|---|
| month | |||
| 1 | 59405722.0 | 61930286.0 | 23598230.0 |
| 2 | 57345684.0 | 60428859.0 | 24446345.0 |
| 3 | 72530252.0 | 32995003.0 | 38050060.0 |
| 4 | 70518994.0 | 3322548.0 | 41826159.0 |
| 5 | 74617773.0 | 7244733.0 | NaN |
Pandas offers other plot types via the kind parameter, so we specify kind='bar' when calling the plot() method. Then, we further format the visualization using the Axes object returned by the plot() method:
import calendar
from matplotlib import ticker
ax = plot_data.plot(
kind='bar', rot=0, xlabel='', ylabel='travelers',
figsize=(8, 1.5), title='TSA Monthly Traveler Throughput'
)
# use month abbreviations for the ticks on the x-axis
ax.set_xticklabels(calendar.month_abbr[1:])
# show y-axis labels in millions instead of scientific notation
ax.yaxis.set_major_formatter(ticker.EngFormatter())
# customize the legend
ax.legend(title='', loc='center', bbox_to_anchor=(0.5, -0.3), ncols=3, frameon=False)
<matplotlib.legend.Legend at 0x119675590>
Some additional things to keep in mind:
- Matplotlib's
tickermodule provides functionality for customizing both the tick labels and locations – check out the documentation for more information. - Pandas supports horizontal and stacked bars as well; this article shows how to make stacked horizontal bars using a pivot table.
- The
plot()method takes a lot of parameters, many of which get passed down to Matplotlib; however, sometimes we need to use Matplotlib calls directly.
Plotting distributions¶
Let's now compare the distribution of daily TSA traveler throughput across years. We will create a subplot for each year with both a histogram and a kernel density estimate (KDE) of the distribution. Pandas has generated the Figure and Axes objects for both examples so far, but we can build custom layouts by creating them ourselves with Matplotlib using the plt.subplots() function. First, we will need to import the pyplot module:
import matplotlib.pyplot as plt
While pandas lets us specify that we want subplots and their layout (with the subplots and layout parameters, respectively), using Matplotlib to create the subplots directly gives us additional flexibility:
# define the subplot layout
fig, axes = plt.subplots(3, 1, sharex=True, sharey=True, figsize=(6, 4))
for year, ax in zip(tsa_melted_holiday_travel.year.unique(), axes):
plot_data = tsa_melted_holiday_travel.loc[str(year)].travelers
plot_data.plot(kind='hist', legend=False, density=True, alpha=0.8, ax=ax)
plot_data.plot(kind='kde', legend=False, color='blue', ax=ax)
ax.set(title=f'{year} TSA Traveler Throughput', xlabel='travelers')
fig.tight_layout() # handle overlaps
Tip: If you're new to the zip() function, check out this article.
Solution¶
We start by handling imports:
from matplotlib import ticker
import pandas as pd
Next, we write a function that will read in the data, use the pivot() method to reshape it, and then use the plot() method to generate the box plot:
def ex1():
df = pd.read_csv('../data/tsa_melted_holiday_travel.csv')
plot_data = df.pivot(columns='year', values='travelers')
ax = plot_data.plot(kind='box')
ax.set(xlabel='year', ylabel='travelers', title='TSA Traveler Throughput')
ax.yaxis.set_major_formatter(ticker.EngFormatter())
return ax
Finally, we call our function:
ex1()
<Axes: title={'center': 'TSA Traveler Throughput'}, xlabel='year', ylabel='travelers'>
Learning Path¶
- Why is data visualization necessary?
- Plotting with pandas
- Plotting with Seaborn
- Customizing plots with Matplotlib
Plotting with Seaborn¶
The Seaborn library provides the means to easily visualize long-format data without first pivoting it. In addition, it also offers some additional plot types – once again building on top of Matplotlib. Here, we will look at a few examples of visualizations we can create with Seaborn.
Visualizing long-format data¶
With Seaborn, we can specify plot colors according to values of a column with the hue parameter. When working with functions that generate subplots, we can also specify how to split the subplots by values of a long-format column with the col and row parameters. Here, we revisit the comparison of the distribution of TSA traveler throughput across years:
import seaborn as sns
sns.displot(
data=tsa_melted_holiday_travel, x='travelers', col='year', kde=True, height=2.5
)
<seaborn.axisgrid.FacetGrid at 0x12a01a660>
Heatmaps¶
We can also use Seaborn to visualize pivot tables as heatmaps:
data = tsa_melted_holiday_travel['2019':'2021-04']\
.assign(month=lambda x: x.index.month)\
.pivot_table(index='month', columns='year', values='travelers', aggfunc='sum')
data
| year | 2019 | 2020 | 2021 |
|---|---|---|---|
| month | |||
| 1 | 59405722.0 | 61930286.0 | 23598230.0 |
| 2 | 57345684.0 | 60428859.0 | 24446345.0 |
| 3 | 72530252.0 | 32995003.0 | 38050060.0 |
| 4 | 70518994.0 | 3322548.0 | 41826159.0 |
| 5 | 74617773.0 | 7244733.0 | NaN |
| 6 | 76619900.0 | 14481802.0 | NaN |
| 7 | 79511968.0 | 20740781.0 | NaN |
| 8 | 74776010.0 | 21708071.0 | NaN |
| 9 | 66531258.0 | 21488263.0 | NaN |
| 10 | 72096495.0 | 25636496.0 | NaN |
| 11 | 68787654.0 | 25512987.0 | NaN |
| 12 | 70219363.0 | 26391765.0 | NaN |
ax = sns.heatmap(data=data / 1e6, cmap='Blues', annot=True, fmt='.1f')
_ = ax.set_yticklabels(calendar.month_abbr[1:], rotation=0)
_ = ax.set_title('Total TSA Traveler Throughput (in millions)')
Tip: Reference the Matplotlib documentation for more information on colormaps and named colors.
We're moving on from Seaborn now, but there is a lot more available in the API. Be sure to check out the following at a minimum:
- pairwise plots with
pairplot() - categorical scatter plots with
swarmplot() - joint distribution plots with
jointplot() - FacetGrids for custom layouts with any plot type
Solution¶
We start by reading in the data and handling imports:
import calendar
from matplotlib import ticker
import pandas as pd
import seaborn as sns
Next, we write a function that will read in the data, create a pivot table, and visualize it as a heatmap:
def ex2():
df = pd.read_csv(
'../data/tsa_melted_holiday_travel.csv',
parse_dates=True, index_col='date'
)
plot_data = df.loc['2019'].assign(
day_of_week=lambda x: x.index.dayofweek, month=lambda x: x.index.month
).pivot_table(
index='day_of_week', columns='month', values='travelers', aggfunc='median'
)
ax = sns.heatmap(data=plot_data / 1e6, annot=True, fmt='.1f', cmap='Blues')
ax.set_xticklabels(calendar.month_abbr[1:])
ax.set_yticklabels(calendar.day_abbr, rotation=0)
ax.set_title('2019 TSA Median Traveler Throughput\n(in millions)')
return ax
Finally, we call our function:
ex2()
<Axes: title={'center': '2019 TSA Median Traveler Throughput\n(in millions)'}, xlabel='month', ylabel='day_of_week'>
Learning Path¶
- Why is data visualization necessary?
- Plotting with pandas
- Plotting with Seaborn
- Customizing plots with Matplotlib
Customizing plots with Matplotlib¶
In this final section, we will discuss how to use Matplotlib to customize plots. Since there is a lot of functionality available, we will only be covering how to add shaded regions and annotations here, but be sure to check out the documentation for more.
Adding shaded regions¶
When looking at a plot of TSA traveler throughput over time, it's helpful to indicate periods during which there was holiday travel. We can do so with the axvspan() method:
plot_data = tsa_melted_holiday_travel['2019-05':'2019-11']
ax = plot_data.travelers.plot(
title='TSA Traveler Throughput', ylabel='travelers', figsize=(9, 2)
)
ax.yaxis.set_major_formatter(ticker.EngFormatter())
# collect the holiday ranges (start and end dates)
holiday_ranges = plot_data.dropna().reset_index()\
.groupby('holiday').agg({'date': ['min', 'max']})
# create shaded regions for each holiday in the plot
for start_date, end_date in holiday_ranges.to_numpy():
ax.axvspan(start_date, end_date, color='gray', alpha=0.2)
Tip: Use axhspan() for horizontally shaded regions and axvline() / axhline() for vertical/horizontal reference lines.
Adding annotations¶
We can use the annotate() method to add annotations to the plot. Here, we point out the day in 2019 with the highest TSA traveler throughput, which was the day after Thanksgiving:
plot_data = tsa_melted_holiday_travel.loc['2019']
ax = plot_data.travelers.plot(
title='TSA Traveler Throughput', ylabel='travelers', figsize=(9, 2)
)
ax.yaxis.set_major_formatter(ticker.EngFormatter())
# highest throughput
max_throughput_date = plot_data.travelers.idxmax()
max_throughput = plot_data.travelers.max()
_ = ax.annotate(
f'{max_throughput_date:%b %d}\n({max_throughput / 1e6:.2f} M)',
xy=(max_throughput_date, max_throughput),
xytext=(max_throughput_date - pd.Timedelta(days=25), max_throughput * 0.92),
arrowprops={'arrowstyle': '->'}, ha='center'
)
Some things to keep in mind:
- We used
Axesmethods to customize our plots (i.e., an object-oriented approach), but thepyplotmodule provides equivalent functions (i.e., a functional approach) for adding shaded regions, reference lines, annotations, etc. – although the function names might be slightly different than theirAxesmethod counterparts (e.g.,Axes.set_xlabel()vs.plt.xlabel()). - In general, try to stick to either the object-oriented or functional approach rather than mixing the two. However, be careful when working with subplots –
pyplotfunctions will only affect the last subplot. - The anatomy of a figure diagram in the Matplotlib documentation is a great resource for identifying which objects you will need to access for plot customizations.
For more on data visualization in Python, including animations and interactive plots, check out my Beyond the Basics: Data Visualization in Python workshop.
Exercise 3.3¶
Annotate the medians in the box plot from Exercise 3.1. Hint: The x coordinates will be 1, 2, and 3 for 2019, 2020, and 2021, respectively. Alternatively, to avoid hardcoding values, you can use the Axes.get_xticklabels() method, in which case you should look at the documentation for the Text class.¶
Solution¶
First, we modify the return statement from the solution to Exercise 3.1 to also give us the data:
from matplotlib import ticker
import pandas as pd
def ex1():
df = pd.read_csv('../data/tsa_melted_holiday_travel.csv')
plot_data = df.pivot(columns='year', values='travelers')
ax = plot_data.plot(kind='box')
ax.set(xlabel='year', ylabel='travelers', title='TSA Traveler Throughput')
ax.yaxis.set_major_formatter(ticker.EngFormatter())
return ax, plot_data
Now, we can build upon ex1() in a new function:
def ex3():
ax, plot_data = ex1()
# add annotations
medians = plot_data.median()
for tick_label in ax.get_xticklabels():
median = medians[int(tick_label.get_text())]
ax.annotate(
f'{median / 1e6:.1f} M',
xy=(tick_label.get_position()[0], median),
ha='center', va='bottom'
)
return ax
Calling our function returns an annotated version of the plot from Exercise 3.1:
ex3()
<Axes: title={'center': 'TSA Traveler Throughput'}, xlabel='year', ylabel='travelers'>
Section 3 Complete 🎉¶

Section 4: Hands-On Data Analysis Lab¶
We will practice all that you’ve learned in a hands-on lab. This section features a set of analysis tasks that provide opportunities to apply the material from the previous sections.

Important notes:¶
- You don't have to answer all the questions to submit.
- There are likely more questions than you will have time to answer, so be sure to submit before the session ends.
Section 4 Complete 🎉¶

Related content¶
All examples herein were developed exclusively for this workshop – check out Hands-On Data Analysis with Pandas and my Beyond the Basics: Data Visualization in Python workshop for more.
Thank you!¶
I hope you enjoyed the session. You can follow my work on these platforms:










